Examples¶
The Examples directory contains examples of data sets on which ProCARs can be run. You will find two different examples: a fictive data set, and a real data set.
Fictive_data¶
Data set description¶
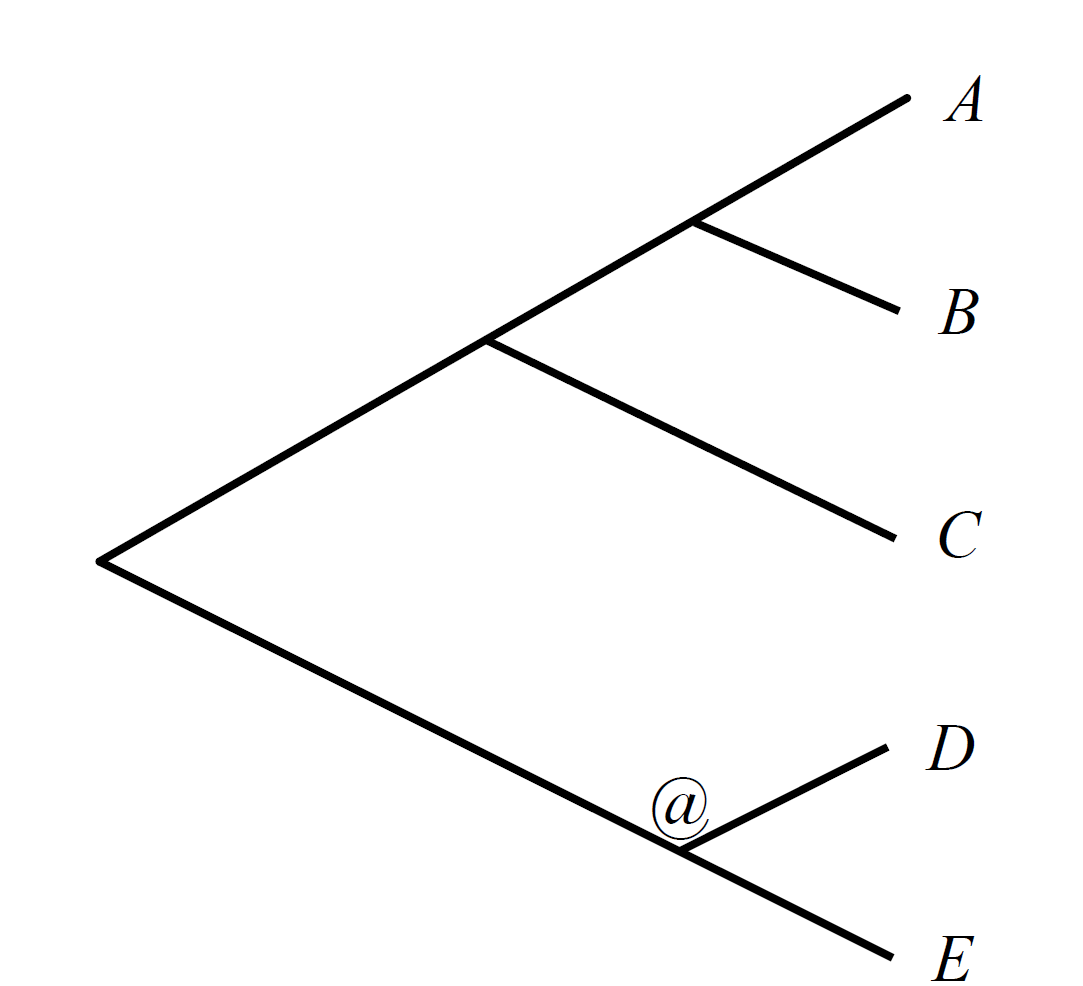
This data set is composed of 5 genomes (A, B, C, D and E) with 8 synteny blocks. It corresponds to the following phylogeny, with the ancestor to reconstruct marked with an @:
You will find in this directory a species tree called tree.ph (corresponding to the tree here-above), and a block file called orthology_blocks.txt. This data set corresponds to the example given in the paper’s method part.
Run data set¶
To run this data set from the Examples directory (if you previously installed ProCARs):
procars_main --tf=Fictive_data/tree.ph --bf=Fictive_data/orthology_blocks.txt -r Fictive_data/results
If you did not install ProCARs, run:
PYTHONPATH+=.. python ../bin/procars_main --tf=Fictive_data/tree.ph --bf=Fictive_data/orthology_blocks.txt -r Fictive_data/results
Results¶
With this dataset, ProCARs runs in 3 steps:
- step1 founds:
- a set of 1 fully conserved and 4 partly conserved non-conflicting adjacencies, which are added to the PQtree: see Fictive_data/results/procars_steps/S_Adjacencies_ancestor_1.txt and Fictive_data/results/procars_steps/S_PQtree_ancestor_1.txt.
- a set of 2 discarded adjacencies: see Fictive_data/results/procars_steps/S_Adjacencies_ancestor_1.txt_discarded.txt
- step2 founds:
- an empty set of non-conflicting adjacencies
- a set of 2 discarded adjacencies (the sames as in step1): see Fictive_data/results/procars_steps/S_Adjacencies_ancestor_2.txt_discarded_used.txt
- Hence, it runs step b) (resolve conflicts), using the set of discarded adjacencies, and retains 1 adjacency, giving the PQtree Fictive_data/results/procars_steps/S_PQtree_ancestor_2.txt
- step3 founds:
- empty sets of non-conflicting and conflicting adjacencies
- Hence, it runs step c) (find DCJ-reliable adjacencies), but does not find any adjacency to add.
The output file is hence Fictive_data/results/ProCars_PQtree.txt, which is the same as Fictive_data/results/procars_steps/S_PQtree_ancestor_2.txt. The ancestor genome contains 2 CARs with 4 blocks each.
Boreoeutherian_ancestor¶
Data set description¶
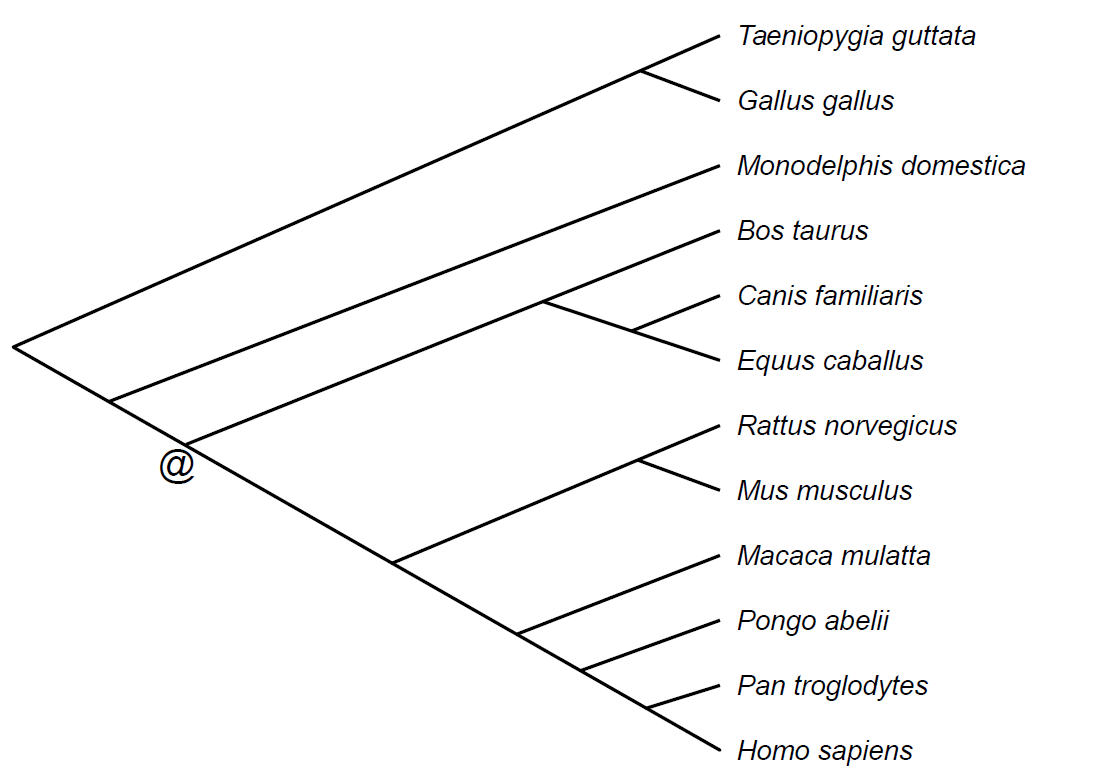
This data set is used to compute a set of CARs for the boreoeutherian ancestral genome using the gene orders of twelve amniote genomes.
We chose those twelve genomes assembled and present in a Pecan multiple alignement of 20 amniote genomes available in the release 73 of the Ensembl Compara database. The phylogenetic tree (tree_boreoeutherian_ancestor.ph) was obtained from the National Center for Biotechnology Information taxonomy, and is the following:
We constructed a set of synteny blocks (orthology_blocks.txt) using the multiple alignements as seeds. This resulted in a set of 12 genomes made of 689 blocks for species Homo sapiens, Pan troglodytes, Pongo abelii, Macaca mulatta, Mus musculus, Rattus norvegicus, Equus caballus, Canis familiaris, Bos taurus, Monodelphis domestica, Gallus gallus and Taeniopygia guttata.
Run data set¶
To run this data set from the Examples directory (if you previously installed ProCARs):
procars_main --tf=Boreoeutherian_ancestor/tree_boreoeutherian_ancestor.ph --bf=Boreoeutherian_ancestor/orthology_blocks.txt -r Boreoeutherian_ancestor/results
If you did not install ProCARs, run:
PYTHONPATH+=.. python ../bin/procars_main --tf=Boreoeutherian_ancestor/tree_boreoeutherian_ancestor.ph --bf=Boreoeutherian_ancestor/orthology_blocks.txt -r Boreoeutherian_ancestor/results
Results¶
With this dataset, ProCARs runs in 5 steps:
- step1:
- a set of 498 fully conserved and 149 partly conserved non-conflicting adjacencies which are added to the PQtree: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_1.txt and Boreoeutherian_ancestor/results/procars_steps/S_PQtree_ancestor_1.txt.
- a set of 11 discarded conflicting adjacencies: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_1.txt_discarded.txt
- step2:
- a set of 10 partly conserved non-conflicting adjacencies which are added to the PQtree: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_2.txt and Boreoeutherian_ancestor/results/procars_steps/S_PQtree_ancestor_2.txt.
- a set of 6 discarded conflicting adjacencies: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_2.txt_discarded.txt
- step3:
- an empty set of non-conflicting adjacencies.
- a set of 6 discarded conflicting adjacencies: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_3.txt_discarded_used.txt
- runs step b) (resolve conflicts), and retains 3 adjacencies, added to the PQtree: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_3.txt and Boreoeutherian_ancestor/results/procars_steps/S_PQtree_ancestor_3.txt.
- step4:
- an empty set of non-conflicting adjacencies.
- an empty set of conflicting adjacencies.
- runs step c) (Find DCJ-reliable adjacencies), and retains 3 DCJ-reliable adjacencies, added to the PQtree: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_4.txt and Boreoeutherian_ancestor/results/procars_steps/S_PQtree_ancestor_4.txt.
- step5:
- a set of 2 partly conserved non-conflicting adjacencies which are added to the PQtree: see Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_5.txt and Boreoeutherian_ancestor/results/procars_steps/S_PQtree_ancestor_5.txt.
- an empty set of conflicting adjacencies.
In step6, all sets of adjacencies are empty, and no DCJ-reliable adjacency is found: the algorithm stops. The output is Boreoeutherian_ancestor/results/ProCars_PQtree.txt, which is the same as (Boreoeutherian_ancestor/results/procars_steps/S_Adjacencies_ancestor_5.txt). ProCARs found 25 CARs for this Boreoeutherian ancestor’s genome, with a number of blocks per ACR ranging from 2 to 68.