What is TFM-Explorer ?
TFM-Explorer (Transcription Factor Matrix Explorer) is a program for analysing regulatory regions in eukaryotic genomes.
It takes a set of coregulated gene sequences, and searches for locally overrepresented transcription factor binding sites.
The algorithm proceeds in two steps:
- scans sequences for detecting all potential transcription factor binding sites, using weight matrices from JASPAR or TRANSFAC (disclaimer)
- extracts significant clusters (region of the input sequences associated with a factor) by calculating a score function.
The method takes advantage of spatial conservation in the sequences and
supports multiple species.
TFM-Explorer allows also to visualize the result clusters and their putative transcription factor binding sites on the input sequences, and to search for cis-regulatory modules.
Example
>Human target genes NM_000040 NM_000043 NM_000073 NM_000074 NM_000118 NM_000201 NM_000367 ...scanned from 2000bp upstream the transcription start site (TSS) until the TSS
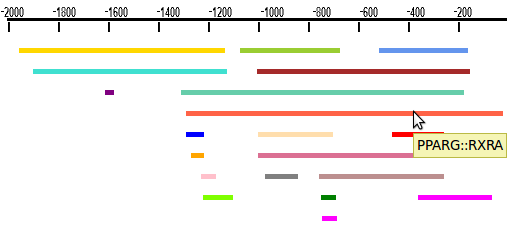
Availability
![]() You can use TFM-Explorer via the web interface.
You can use TFM-Explorer via the web interface.
![]() You can also download a command line version here.
You can also download a command line version here.
References
TFM-Explorer: mining cis-regulatory regions in genomes
Laurie Tonon, Helene Touzet and Jean-Stephane Varre
Nucleic Acids Research 2010
Predicting transcription factor binding sites using local over-representation and comparative genomics
Matthieu Defrance, Helene Touzet
BMC Bioinformatics 2006